In this tutorial, we will use the fpp3 library in R to manipulate and plot time series data. fpp3 loads other useful packages such as: dplyr, tidyr, lubridate, and ggplot2.
library(fpp3)
We will start with a simple example and then work with a more complicated one.
1. A simple time series example
1.1. Create the data
First, we will simulate some data (after setting the seed for reproducibility):
set.seed(1) cancer <- data.frame( year = rep(2023, 12), month = 1:12, new_cases = sample(1:50, 12, replace = TRUE) ) cancer # year month new_cases #1 2023 1 4 #2 2023 2 39 #3 2023 3 1 #4 2023 4 34 #5 2023 5 23 #6 2023 6 43 #7 2023 7 14 #8 2023 8 18 #9 2023 9 33 #10 2023 10 21 #11 2023 11 21 #12 2023 12 42
Next, we will combine the variables year and month into 1 date variable:
# make_yearmonth() is available from fpp3 cancer <- cancer |> mutate(date = make_yearmonth(year, month)) |> select(-c(year, month)) # remove year and month cancer # new_cases date #1 4 2023 Jan #2 39 2023 Feb #3 1 2023 Mar #4 34 2023 Apr #5 23 2023 May #6 43 2023 Jun #7 14 2023 Jul #8 18 2023 Aug #9 33 2023 Sep #10 21 2023 Oct #11 21 2023 Nov #12 42 2023 Dec
1.2. Create a time series object
A time series table (i.e. a tsibble object) will make it easier for us to manipulate, analyze, and plot the time series data.
The function as_tsibble() requires us to specify the time index variable, which in this case is date:
cancer_ts <- as_tsibble(cancer,
index = date)
cancer_ts
## A tsibble: 12 x 2 [1M]
# new_cases date
# <int> <mth>
# 1 4 2023 Jan
# 2 39 2023 Feb
# 3 1 2023 Mar
# 4 34 2023 Apr
# 5 23 2023 May
# 6 43 2023 Jun
# 7 14 2023 Jul
# 8 18 2023 Aug
# 9 33 2023 Sep
#10 21 2023 Oct
#11 21 2023 Nov
#12 42 2023 Dec
The first line in the output (# A tsibble: 12 x 2 [1M]) is telling us that we have a time series table of 12 observations and 2 variables (12 × 2) which represent monthly data ([1M]).
1.3. Plot time series
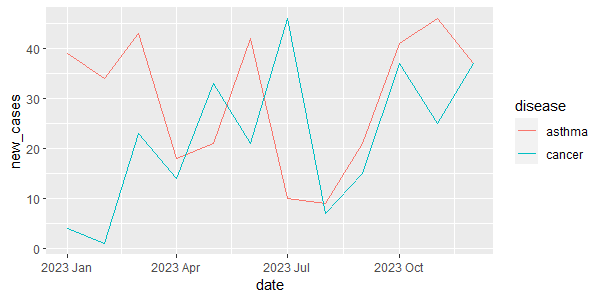
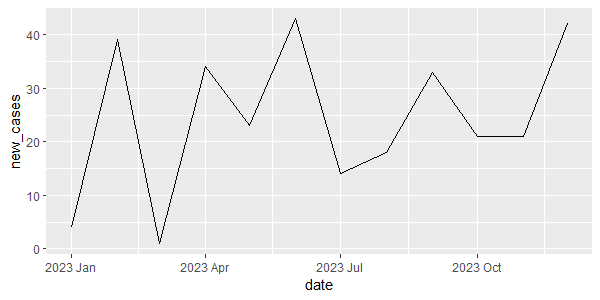
# 1st option cancer_ts |> ggplot(aes(x = date, y = new_cases)) + geom_line() # alternatively, we can use autoplot autoplot(cancer_ts, new_cases)
Output:
2. A more complicated time series example
2.1. Create the data
In this second example, we will simulate time series data for 2 diseases: cancer and asthma.
set.seed(1)
diseases <- data.frame(
year = rep(2023, 24), # 2 years
month = rep(1:12, each = 2), # 2 years
disease = rep(c("cancer", "asthma"), 12),
new_cases = sample(1:50, 24, replace = TRUE)
)
diseases
# year month disease new_cases
#1 2023 1 cancer 4
#2 2023 1 asthma 39
#3 2023 2 cancer 1
#4 2023 2 asthma 34
#5 2023 3 cancer 23
#6 2023 3 asthma 43
#7 2023 4 cancer 14
#8 2023 4 asthma 18
#9 2023 5 cancer 33
#10 2023 5 asthma 21
#11 2023 6 cancer 21
#12 2023 6 asthma 42
#13 2023 7 cancer 46
#14 2023 7 asthma 10
#15 2023 8 cancer 7
#16 2023 8 asthma 9
#17 2023 9 cancer 15
#18 2023 9 asthma 21
#19 2023 10 cancer 37
#20 2023 10 asthma 41
#21 2023 11 cancer 25
#22 2023 11 asthma 46
#23 2023 12 cancer 37
#24 2023 12 asthma 37
Next, we will combine the variables year and month into 1 date variable:
diseases <- diseases |> mutate(date = make_yearmonth(year, month)) |> select(-c(year, month)) # remove year and month diseases # disease new_cases date #1 cancer 4 2023 Jan #2 asthma 39 2023 Jan #3 cancer 1 2023 Feb #4 asthma 34 2023 Feb #5 cancer 23 2023 Mar #6 asthma 43 2023 Mar #7 cancer 14 2023 Apr #8 asthma 18 2023 Apr #9 cancer 33 2023 May #10 asthma 21 2023 May #11 cancer 21 2023 Jun #12 asthma 42 2023 Jun #13 cancer 46 2023 Jul #14 asthma 10 2023 Jul #15 cancer 7 2023 Aug #16 asthma 9 2023 Aug #17 cancer 15 2023 Sep #18 asthma 21 2023 Sep #19 cancer 37 2023 Oct #20 asthma 41 2023 Oct #21 cancer 25 2023 Nov #22 asthma 46 2023 Nov #23 cancer 37 2023 Dec #24 asthma 37 2023 Dec
2.2. Create time series object
Since we have observations for 2 diseases (cancer and asthma), i.e. for each date we have 2 rows, we will need to tell that to R by setting key = disease inside as_tsibble().
diseases_ts <- as_tsibble(diseases,
index = date,
key = disease)
diseases_ts
## A tsibble: 24 x 3 [1M]
## Key: disease [2]
# disease new_cases date
# <chr> <int> <mth>
# 1 asthma 39 2023 Jan
# 2 asthma 34 2023 Feb
# 3 asthma 43 2023 Mar
# 4 asthma 18 2023 Apr
# 5 asthma 21 2023 May
# 6 asthma 42 2023 Jun
# 7 asthma 10 2023 Jul
# 8 asthma 9 2023 Aug
# 9 asthma 21 2023 Sep
#10 asthma 41 2023 Oct
## ℹ 14 more rows
## ℹ Use `print(n = ...)` to see more rows
2.3. Plot time series
# 1st option diseases_ts |> ggplot(aes(x = date, y = new_cases, color = disease)) + geom_line() # alternatively, we can use autoplot() autoplot(diseases_ts, new_cases)
Output: