In this article, we will produce the following plot in R:
1. Load the data
First, we will use the COVID19 package in R to download data for Lebanon. We will limit our analysis to 2 variables: (1) the date and (2) the number of new daily cases.
library(COVID19) # load data library(tidyverse) # manipulate data dat <- tibble(covid19(country = "Lebanon")) |> mutate(new_cases = confirmed - lag(confirmed)) |> select(date, new_cases) dat ## A tibble: 1,162 × 2 # date daily_cases # <date> <int> # 1 2020-01-03 NA # 2 2020-01-04 NA # 3 2020-01-05 NA # 4 2020-01-06 NA # 5 2020-01-07 NA # 6 2020-01-08 NA # 7 2020-01-09 NA # 8 2020-01-10 NA # 9 2020-01-11 NA #10 2020-01-12 NA ## ℹ 1,152 more rows ## ℹ Use `print(n = ...)` to see more rows
2. Count monthly cases
To do that, we group data by year and month, and then we count the cases:
# count new cases each month
dat <- dat |>
group_by(year = year(date),
month = month(date)) |>
count(wt = daily_cases, name = "monthly_cases")
dat
## A tibble: 39 × 3
## Groups: year, month [39]
# year month monthly_cases
# <dbl> <dbl> <int>
# 1 2020 1 0
# 2 2020 2 3
# 3 2020 3 466
# 4 2020 4 255
# 5 2020 5 495
# 6 2020 6 558
# 7 2020 7 2777
# 8 2020 8 12753
# 9 2020 9 22326
#10 2020 10 41594
## ℹ 29 more rows
## ℹ Use `print(n = ...)` to see more rows
3. Transform to time series table (tsibble)
Next, we transform the tibble obejct into a tsibble using the fpp3 package, and we specify that we want monthly data to be the index of the table:
library(fpp3) # use time series tables # transform to time series dat <- dat |> unite(date, year, month) |> mutate(date = yearmonth(date)) |> as_tsibble(index = date) dat ## A tsibble: 39 x 2 [1M] # date monthly_cases # <mth> <int> # 1 2020 Jan 0 # 2 2020 Feb 3 # 3 2020 Mar 466 # 4 2020 Apr 255 # 5 2020 May 495 # 6 2020 Jun 558 # 7 2020 Jul 2777 # 8 2020 Aug 12753 # 9 2020 Sep 22326 #10 2020 Oct 41594 ## ℹ 29 more rows ## ℹ Use `print(n = ...)` to see more rows
4. Plot the monthly distribution
Now, we can use the function gg_season() and specify that we want to plot the variable called monthly_cases:
df |> gg_season(monthly_cases) + geom_point()
Output:
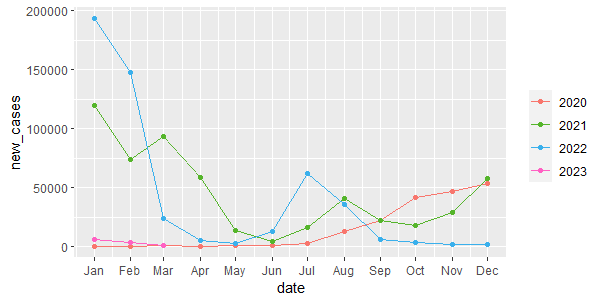
Interpretation: The graph illustrates the seasonal variation of COVID-19 infection rate in Lebanon from 2020 to mid 2023. The infection rate reached its highest levels in the winter months (November to February) and in the summer months (July and August). These periods coincide with longer indoor stays due to cold weather and higher tourism activities respectively. The infection rate declined significantly by the end of 2022.