In this tutorial, we will use the packages ggmap and COVID19 to create the following plot:
1. Downloading and plotting the map
Since we are going to plot the Mediterranean region only, we first need to specify its borders. A simple Google search shows that these are roughly:
- Between -10 and 40 in longitude.
- Between 18 and 50 in latitude.
library(ggmap)
library(ggthemes) # for theme_map()
# Mediterranean region borders
area <- c(left = -10,
right = 40,
bottom = 18,
top = 50)
# download the map
# zoom=5 avoids downloading a lot of data about this region
med_map <- get_stamenmap(bbox = area,
zoom = 5)
# plot the map
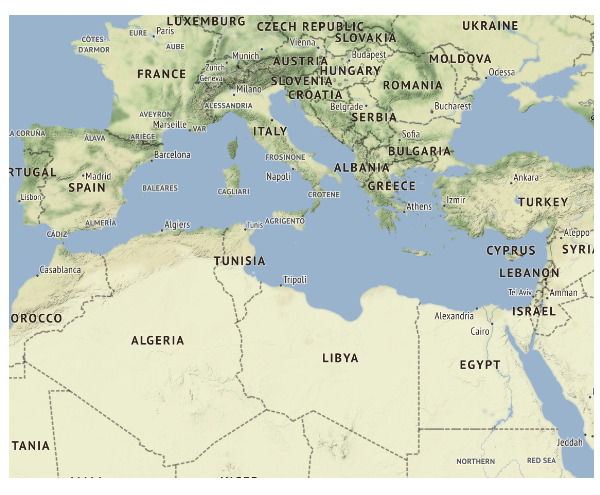
ggmap(med_map) +
theme_map()
Output:
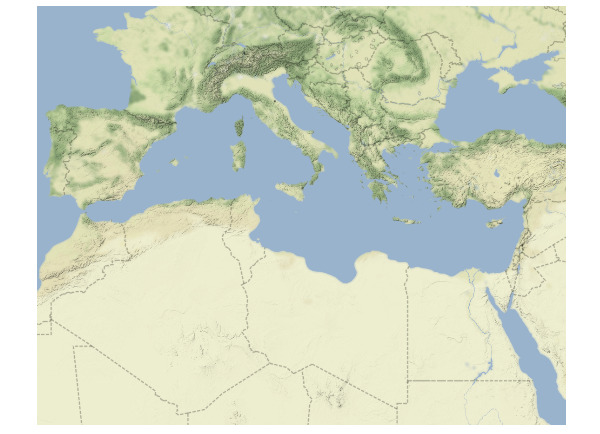
Since we don’t want country labels in our map, we will set maptype = "terrain-background" inside the get_stamenmap() function:
# download the map
med_map <- get_stamenmap(bbox = area,
zoom = 5,
maptype = "terrain-background")
# plot the map
ggmap(med_map) +
theme_map()
Output:
2. Plotting COVID-19 deaths data on the map
Next, we will load the COVID19 package in R to get the deaths statistics for COVID-19. We will also use the tidyverse package for some data manipulation.
library(COVID19) library(tidyverse) df <- covid19()
The dataset has a lot of variables, but we are only interested in 3: longitude, latitude, and deaths. We will also limit the data to our region of interest:
region_df <- df |> select(date, longitude, latitude, deaths) |> filter(!is.na(deaths)) |> filter(longitude > -10 & longitude < 40) |> filter(latitude > 18 & latitude < 50) head(region_df) # date longitude latitude deaths #1 2020-03-18 29 47 1 #2 2020-03-19 29 47 1 #3 2020-03-20 29 47 1 #4 2020-03-21 29 47 1 #5 2020-03-22 29 47 1 #6 2020-03-23 29 47 1
Since the variable deaths represents the cumulative number of deaths, we only need the last number (corresponding to the last date) for each combination of longitude and latitude:
deaths_df <- region_df |> group_by(longitude, latitude) |> slice_tail(n = 1) head(deaths_df) ## A tibble: 6 x 4 ## Groups: longitude, latitude [6] # date longitude latitude deaths # <date> <dbl> <dbl> <int> #1 2022-05-10 -8.22 39.4 23004 #2 2023-03-09 -7.09 31.8 16296 #3 2023-03-09 -5.35 36.1 111 #4 2023-03-09 -4 40 119479 #5 2023-03-09 1.52 42.5 165 #6 2022-10-06 1.66 28.0 6881
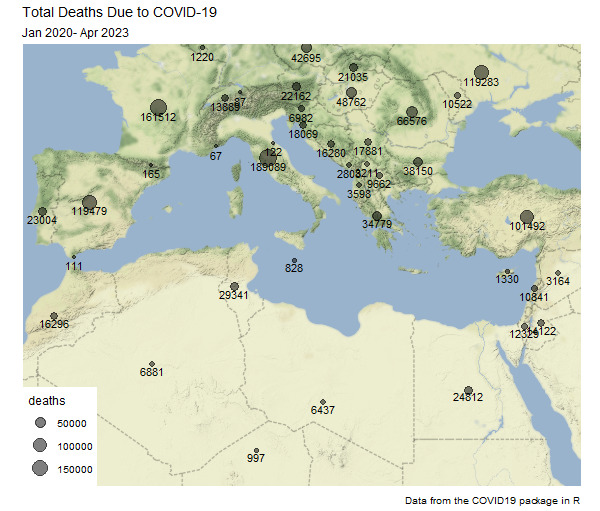
Adding these data to the map:
ggmap(med_map) +
theme_map() +
geom_point(aes(x = longitude, y = latitude, size = deaths),
alpha = 0.5,
data = deaths_df) +
geom_text(aes(x = longitude, y = latitude, label = deaths),
size = 3,
nudge_y = -0.5,
data = deaths_df) +
labs(title = "Total Deaths Due to COVID-19",
subtitle = "Jan 2020- Apr 2023",
caption = "Data from the COVID19 package in R")
Output:
