When running a linear regression model:
Y = β0 + β1 × X1 + β2 × X2 + ε
One way of determining if the independent variables X1 and X2 were useful in predicting Y is to calculate the coefficient of determination R2.
R2 measures the proportion of variability in Y that can be explained by X1 and X2.
For example, an R2 of 0.3 means that the linear regression model (with predictors X1 and X2) can explain 30% of the variability in Y.
This also means that 70% of the variability in Y cannot be explained by X1 and X2. This quantity, 1 – R2, is referred to as the coefficient of non-determination or coefficient of alienation.
The coefficient of alienation (1 – R2) is a measure of the variability in the outcome Y that remains unexplained by the linear regression model. Since R2 is a number between 0 and 1, the coefficient of alienation will also be between 0 and 1.
What is a good value for the coefficient of alienation/non-determination?
When we model an outcome Y, we would like to explain as much of its variability as we can. Therefore the lower the coefficient of alienation the better.
However, in the social sciences, epidemiology and health research in general, it is very hard to explain an outcome fully using some model. So in these fields, regression models with high coefficient of alienation are seen all the time.
Here’s an example:
A study tried to model systolic blood pressure in men using age and percentage of total body fat as predictors:
SBP = Age + Fat percentage
The R2 of this model was 0.183.
Therefore, 81.7% of the variability in systolic blood pressure was left unexplained by the linear regression (which sounds reasonable as blood pressure is a more complex variable to be totally predicted by age and body fat only).
Tolerance
Consider a linear regression model with 3 predictors (X1, X2 and X3), ideally these predictors:
- Should be highly correlated with Y
- Shouldn’t be correlated with each other — no multicollinearity
In order to determine if these predictors are related to each other, we can model each one using all the others by running the following linear regression models:
X1 = β0 + β1 × X2 + β2 × X3
X2 = β0 + β1 × X1 + β2 × X3
X3 = β0 + β1 × X1 + β2 × X2
Then we calculate the coefficient of determination R2 for each of these models.
If the predictors are not related to each other, R2 for each of these models will be small — So the coefficient of alienation will be large.
In this context the coefficient of alienation (1 – R2) is referred to as tolerance, and we want it to be as large as possible in order to avoid multicollinearity.
You may be familiar with another measure of multicollinearity, the variance inflation factor VIF, which is the inverse of tolerance:
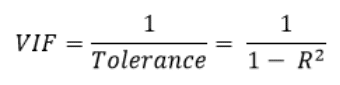
As a rule of thumb, a tolerance level below 0.1 (which corresponds to a VIF > 10) signals a collinearity problem [source: Regression Methods in Biostatistics, Vittinghoff et al.], in this case consider removing 1 of the variables that may contain redundant information.
That being said, choosing a VIF threshold to detect collinearity can get a little bit more complex than that, with different references recommending different thresholds. For more information on this topic and on how to interpret a VIF value, I recommend my other article: What is an Acceptable Value for VIF?